Show code cell source
from IPython.display import YouTubeVideo
YouTubeVideo("TEkLFmqi-Bw")
PyData NYC ‘23#
E-graphs in Python with egglog
Saul Shanabrook
Aims:#
Faithful bindings for
egglogrust library.“Pythonic” interface, using standard type definitions.
Usable as base for optimizing/translating expressions for data science libraries in Python
What is an e-graph?#
E-graphs are these super wonderful data structures for managing equality and equivalence information. They are traditionally used inside of constraint solvers and automated theorem provers to implement congruence closure, an efficient algorithm for equational reasoning—but they can also be used to implement rewrite systems.
Define types and functions/operators
Define rewrite rules
Add expressions to graph
Run rewrite rules on expressions until saturated (addtional applications have no effect)
Extract out lowest cost expression
Example#
from __future__ import annotations
from egglog import *
egraph = EGraph()
@egraph.class_
class NDArray(Expr):
def __init__(self, i: i64Like) -> None:
...
def __add__(self, other: NDArray) -> NDArray:
...
def __mul__(self, other: NDArray) -> NDArray:
...
@egraph.function(cost=2)
def arange(i: i64Like) -> NDArray:
...
# Register rewrite rule that asserts for all values x of type NDArray
# x + x = x * 2
x = var("x", NDArray)
egraph.register(rewrite(x + x).to(x * NDArray(2)))
res = arange(10) + arange(10)
egraph.register(res)
egraph.saturate()
egraph.extract(res)
arange(10) * NDArray(2)
Example with Scikit-learn#
Optimize Scikit-learn function with Numba by building an e-graph that implements the Array API.
from sklearn import config_context
from sklearn.datasets import make_classification
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
def run_lda(x, y):
with config_context(array_api_dispatch=True):
lda = LinearDiscriminantAnalysis()
return lda.fit(x, y).transform(x)
X_np, y_np = make_classification(random_state=0, n_samples=1000000)
run_lda(X_np, y_np)
array([[ 0.64233002],
[ 0.63661245],
[-1.603293 ],
...,
[-1.1506433 ],
[ 0.71687176],
[-1.51119579]])
from egglog.exp.array_api import *
from egglog.exp.array_api_jit import jit
@jit
def optimized_fn(X, y):
# Add metadata about input shapes and dtypes, so that abstract array
# can pass scikit-learn runtime checks
assume_dtype(X, X_np.dtype)
assume_shape(X, X_np.shape)
assume_isfinite(X)
assume_dtype(y, y_np.dtype)
assume_shape(y, y_np.shape)
assume_value_one_of(y, (0, 1))
return run_lda(X, y)
Here is an example of a rewrite rule we used to generate Numba compatible code:
rewrite(
std(x, axis)
).to(
sqrt(mean(square(x - mean(x, axis, keepdims=TRUE)), axis))
)
We can see the optimized expr:
optimized_fn.expr
_NDArray_1 = NDArray.var("X")
assume_dtype(_NDArray_1, DType.float64)
assume_shape(_NDArray_1, TupleInt(Int(1000000)) + TupleInt(Int(20)))
assume_isfinite(_NDArray_1)
_NDArray_2 = NDArray.var("y")
assume_dtype(_NDArray_2, DType.int64)
assume_shape(_NDArray_2, TupleInt(Int(1000000)))
assume_value_one_of(_NDArray_2, TupleValue(Value.int(Int(0))) + TupleValue(Value.int(Int(1))))
_NDArray_3 = astype(
NDArray.vector(TupleValue(sum(_NDArray_2 == NDArray.scalar(Value.int(Int(0)))).to_value()) + TupleValue(sum(_NDArray_2 == NDArray.scalar(Value.int(Int(1)))).to_value())),
DType.float64,
) / NDArray.scalar(Value.float(Float(1000000.0)))
_NDArray_4 = zeros(TupleInt(Int(2)) + TupleInt(Int(20)), OptionalDType.some(DType.float64), OptionalDevice.some(_NDArray_1.device))
_MultiAxisIndexKey_1 = MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(Slice()))
_IndexKey_1 = IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.int(Int(0))) + _MultiAxisIndexKey_1)
_NDArray_5 = _NDArray_1[ndarray_index(_NDArray_2 == NDArray.scalar(Value.int(Int(0))))]
_OptionalIntOrTuple_1 = OptionalIntOrTuple.some(IntOrTuple.int(Int(0)))
_NDArray_4[_IndexKey_1] = sum(_NDArray_5, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_5.shape[Int(0)]))
_IndexKey_2 = IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.int(Int(1))) + _MultiAxisIndexKey_1)
_NDArray_6 = _NDArray_1[ndarray_index(_NDArray_2 == NDArray.scalar(Value.int(Int(1))))]
_NDArray_4[_IndexKey_2] = sum(_NDArray_6, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_6.shape[Int(0)]))
_NDArray_7 = concat(TupleNDArray(_NDArray_5 - _NDArray_4[_IndexKey_1]) + TupleNDArray(_NDArray_6 - _NDArray_4[_IndexKey_2]), OptionalInt.some(Int(0)))
_NDArray_8 = square(_NDArray_7 - expand_dims(sum(_NDArray_7, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_7.shape[Int(0)]))))
_NDArray_9 = sqrt(sum(_NDArray_8, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_8.shape[Int(0)])))
_NDArray_10 = copy(_NDArray_9)
_NDArray_10[ndarray_index(_NDArray_9 == NDArray.scalar(Value.int(Int(0))))] = NDArray.scalar(Value.float(Float(1.0)))
_TupleNDArray_1 = svd(sqrt(NDArray.scalar(Value.float(Float.rational(Rational(1, 999998))))) * (_NDArray_7 / _NDArray_10), FALSE)
_Slice_1 = Slice(OptionalInt.none, OptionalInt.some(sum(astype(_TupleNDArray_1[Int(1)] > NDArray.scalar(Value.float(Float(0.0001))), DType.int32)).to_value().to_int))
_NDArray_11 = (_TupleNDArray_1[Int(2)][IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(_Slice_1)) + _MultiAxisIndexKey_1)] / _NDArray_10).T / _TupleNDArray_1[
Int(1)
][IndexKey.slice(_Slice_1)]
_TupleNDArray_2 = svd(
(sqrt((NDArray.scalar(Value.int(Int(1000000))) * _NDArray_3) * NDArray.scalar(Value.float(Float(1.0)))) * (_NDArray_4 - (_NDArray_3 @ _NDArray_4)).T).T @ _NDArray_11, FALSE
)
(
(_NDArray_1 - (_NDArray_3 @ _NDArray_4))
@ (
_NDArray_11
@ _TupleNDArray_2[Int(2)].T[
IndexKey.multi_axis(
_MultiAxisIndexKey_1
+ MultiAxisIndexKey(
MultiAxisIndexKeyItem.slice(
Slice(
OptionalInt.none,
OptionalInt.some(
sum(astype(_TupleNDArray_2[Int(1)] > (NDArray.scalar(Value.float(Float(0.0001))) * _TupleNDArray_2[Int(1)][IndexKey.int(Int(0))]), DType.int32))
.to_value()
.to_int
),
)
)
)
)
]
)
)[IndexKey.multi_axis(_MultiAxisIndexKey_1 + MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(Slice(OptionalInt.none, OptionalInt.some(Int(1))))))]
And the generated code:
import inspect
print(inspect.getsource(optimized_fn))
def __fn(X, y):
assert X.dtype == np.dtype(np.float64)
assert X.shape == (1000000, 20,)
assert np.all(np.isfinite(X))
assert y.dtype == np.dtype(np.int64)
assert y.shape == (1000000,)
assert set(np.unique(y)) == set((0, 1,))
_0 = y == np.array(0)
_1 = np.sum(_0)
_2 = y == np.array(1)
_3 = np.sum(_2)
_4 = np.array((_1, _3,)).astype(np.dtype(np.float64))
_5 = _4 / np.array(1000000.0)
_6 = np.zeros((2, 20,), dtype=np.dtype(np.float64))
_7 = np.sum(X[_0], axis=0)
_8 = _7 / np.array(X[_0].shape[0])
_6[0, :] = _8
_9 = np.sum(X[_2], axis=0)
_10 = _9 / np.array(X[_2].shape[0])
_6[1, :] = _10
_11 = _5 @ _6
_12 = X - _11
_13 = np.sqrt(np.array(float(1 / 999998)))
_14 = X[_0] - _6[0, :]
_15 = X[_2] - _6[1, :]
_16 = np.concatenate((_14, _15,), axis=0)
_17 = np.sum(_16, axis=0)
_18 = _17 / np.array(_16.shape[0])
_19 = np.expand_dims(_18, 0)
_20 = _16 - _19
_21 = np.square(_20)
_22 = np.sum(_21, axis=0)
_23 = _22 / np.array(_21.shape[0])
_24 = np.sqrt(_23)
_25 = _24 == np.array(0)
_24[_25] = np.array(1.0)
_26 = _16 / _24
_27 = _13 * _26
_28 = np.linalg.svd(_27, full_matrices=False)
_29 = _28[1] > np.array(0.0001)
_30 = _29.astype(np.dtype(np.int32))
_31 = np.sum(_30)
_32 = _28[2][:_31, :] / _24
_33 = _32.T / _28[1][:_31]
_34 = np.array(1000000) * _5
_35 = _34 * np.array(1.0)
_36 = np.sqrt(_35)
_37 = _6 - _11
_38 = _36 * _37.T
_39 = _38.T @ _33
_40 = np.linalg.svd(_39, full_matrices=False)
_41 = np.array(0.0001) * _40[1][0]
_42 = _40[1] > _41
_43 = _42.astype(np.dtype(np.int32))
_44 = np.sum(_43)
_45 = _33 @ _40[2].T[:, :_44]
_46 = _12 @ _45
return _46[:, :1]
As well as the e-graph:
optimized_fn.egraph
import numba
import numpy as np
numba_fn = numba.njit(fastmath=True)(optimized_fn)
assert np.allclose(run_lda(X_np, y_np), numba_fn(X_np, y_np))
/var/folders/xn/05ktz3056kqd9n8frgd6236h0000gn/T/egglog-82012193-cafe-439c-94d4-48ec846ca9e6.py:56: NumbaPerformanceWarning: '@' is faster on contiguous arrays, called on (Array(float64, 2, 'C', False, aligned=True), Array(float64, 2, 'A', False, aligned=True))
_45 = _33 @ _40[2].T[:, :_44]
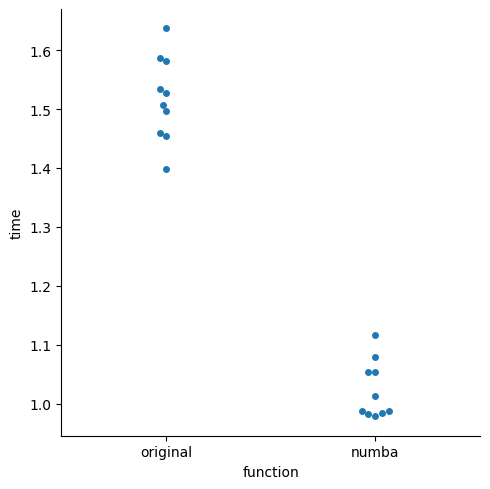
~30% speedup#
on my machine, not a scientific benchmark
import seaborn as sns
df_melt = pd.melt(df, var_name="function", value_name="time")
_ = sns.catplot(data=df_melt, x="function", y="time", kind="swarm")
Conclusions#
egglogis a Python interface to e-graphs, which respects the underlying semantics but provides a Python interface.Flexible enough to represent Array API and translate this back to Python source
If you have a Python library which optimizes/translates expressions, try it out!
Goals
support the ecosystem in collaborating better between libraries, to encourage experimentation and innovation
dont reimplement the world: build on academic programming language research
pip install eggloghttps://github.com/egraphs-good/egglog-python
Say hello: https://egraphs.zulipchat.com/