egglog#
Bringing e-graphs to Python#
Saul Shanabrook

PyData open source needs PL research to handle growing complexity
PL research can have large impact by working with PyData open source
Overview
Why pydata (scientific computing) and PL researchers need each other…
Python open source ecosystem around data, ML, scientific computing needs support of abstractions
PL can use this rich ecosystem to promote its ideas to larger audiences, make them accessible, have large impact, and use the real world problem to drive research goals
How a library for building e-graphs could help them come together
Contributed to…#
Worked at…#
Applying to Ph.D. programs in
programming languages/formal methods.#
My background
Been working in open source python ecosystem 10 years
Slide with logos of projects
different communities, mainting projects
startups, consulting, grants
Applying to PhD programs in PL
My own journey, reflective of needs of community, what that can offer research
mutually beneficial collaboration
First, some history….#
[PYTHON MATRIX-SIG] Time for recap? Fri, 29 Sep 1995
How about separating out the representation and the structure?
I believe I’ve seen C/Fortran matrix packages that made quite good use of this. The representation would be a simple 1-dimensional sequence. You’d normally not see or use this, but it would be there if you needed access to it (e.g. for passing to C/Fortran code).
There’s a simple way to map an index in an N-dim array into an index in the 1-dim representation array (Fortran compilers use it all the time :-).
To make efficient use of this, I propose that slicing and indexing, as long as they return an object of rank >= 1, return an object that points into the same representation sequence.
I propose one additional feature (which I believe I saw in Algol-68; some matrix packages may also use it): add a “stride” to each dimension (including the last). This makes it possible to have slices reference discontiguous parts of the underlying representation, and even to represent a transposed version!
If we do things just right, it may be possible to pass in the sequence to be used as the representation – it could be a Python list, tuple or array (from the array module).
Again, all this can be prototyped in Python!
I would give an example, but I have to run.
–Guido van Rossum guido@CNRI.Reston.VA.US
mixing the representation from the implementation
Pointer, strides, dtype
What is built on top of this?
The term “ecosystem” is often used to describe the modern open-source scientific software. In biology, the term “ecosystem” is defined as a biological community of interacting organisms and their physical environment. Modern open-source scientific software development occurs in a similarly interconnected and interoperable fashion.
Jupyter Meets the Earth: Ecosystem
Why does Python need PL?
It’s in jeopardy now… but to explain how first have to start with history
History of Python open source
interdependent collection of projects
ecosystem
governed independently
Often Multi-stakeholder, from institutions or individuals
depend on one another
users will pull in multiple different projects to achieve their goals, that all work together
This is why its so powerful, someone already built the library and also that the whole ecosystem can function as one big solution
I love working in this space
room for collaboration from diverse individuals
values of transparency, openess, and collective good
Python scientific computing ecosystem#
Original where it took off
NumPy & SciPy
created by Travis Oliphant, who I worked with and serves as a mentor in this pace
Call out to C and Fortran routines
pointer in memory, metadata about shape and dtype
efficient, not have to leave python
Pandas, scikit learn, matplotlib built on top of this
common abstractions, allows users to take array from numpy, pass to scikit learn, then plot with matplotlib
efficient, no memory copies required, becuase all understand common format of numpy pointers at C level, or numpy API at python level
allows projects to be written by diverse people, innovation.
How do we support todays targets?#
I made this four years ago, but gives a sense of some of the complexity
Different from today
changes in hardware
no longer single CPU
multicore
distributed accross many computers
heterogenous compute
GPU
TPU
ASICs
constantly changing as well, not fixed.
Arrays are no longer a pointer to contigious memory and metadata…#
abstractions no longe rhold
pointer to array in memory + shape and type not sufficient as an interface between libraries
If I want to use pytorch deep learning TODO EXAMPLE THAT ISNT POSSIBLE WITH PYTORCH
memory might just be in CPU, might be on a GPU
what if its actually chunked across a number of CPUs?
also want to do whole program optimization, instead of going eagerly back and forth to compute and then to Python after every op, might want to analyze larger chunk of ops together and lower them
How do we manage the new complexity?#
Integrated frameworks funded by companies with 💸?
More complex!
the workflows that manage this are often all encompassing, ala PyTorch, or TF/Jax, or even more so Mojo
Owned by single company!
PyTorch does have open gov, but all core maintainors are at meta
Mojo basically fork the whole language, with lots of $$$$
Users are locked into depending on these stacks, if you want to support the best performance
focus of these companies is more narrow than scientific computing, prioritize their profit centers, of AI
Many libraries are also re-implementing partial Python semantics
Jax, Numba, Tensorflow PyTorch…
to do whole program optimization must deal with parsing Python control flow
Libraries dont work together as well
users end up having to choose one stack… incompatible with others
if they want to change, must rewrite code
if new hardware comes u that isnt covered, must rewrite
for example probabilistic programming, have to choose one
Harder to have smaller isolated libraries that do only their strength
to do this write, you have to basically be a whole compiler
need to support lazy/eager
This is a crisis! We need help! What exactly do we need help with?
If we have tools to manage our complexity, can we have an ecosystem of interrelated DSLs and optimizations?
If we take a step back, we can see the ecosystem as a whole as one big compiler itself
But a ideosyncratic one with lots of mutually incompatible features
If they are part of large compiler, what if they didnt have to re-implement that every time?
by having careful ways of managing abstraction
So what if we lean into the idea of all of these different domains, instead of trying to build a monolothic framework/compiler to rule them all, what if instead we just make it easier for different libraries to write their own piece?
Hardware vendor: write just mapping from low level imperative domain to their hardware
Math array library: write array to hardware
End user: Target the domain they are familiar with
optimizations: could be added seperate
seperate of concerns
Looks much like the goals of MLIR, BUT
Python first
MLIR is imperative first, it says very little about the underlying semantics of any domain
Can we instead have a functional representation? Treat that as the underlying meta-domain?
Whats missing is shared abstractions
this is where it gets to be about people, they are domain experts, building tools for themselves
has been successful so far
but at a place where complexity has grown too much
folks more interested in managing this complexity through abstraction often end up in other places
julia, haskell, dependently typed languages
Why is this relevant to PL?
If we could have a place for these shared abstractions in Python TODO
Could e-graphs help?#

e-graphs
Could they be a foundation we are looking for here?
Overview
How many people understand e-graph? Intuitive understanding?
im newer to this world, just started learning about them in the past year
very grateful to the work of the egg team, Max, Zach, Oliver, at UW and Berkeley for making accessible
Im not an expert
How to use egglog in Python?#
Describe domain with types & functions
Write rewrite rules to express semantics
Add expresions of interest
Run rules
Extract out lowest cost
from __future__ import annotations
from egglog import *
egraph = EGraph()
# 1. Describe domain with types & functions
@egraph.class_
class Num(Expr):
def __init__(self, i: i64Like) -> None:
...
def __add__(self, other: Num) -> Num:
...
@egraph.function(cost=20)
def fib(x: i64Like) -> Num:
...
Working on Python bindings, I will show you some visualize examples
Fib
Create some abstract types & functions
types work with existing linting
Python can use static types
Building DSLs with this feels natural, not a seperate ecosystem, compatible
No actual body, so what if I call it?
# 2. Write rewrite rules to express semantics
@egraph.register
def _fib(a: i64, b: i64):
yield rewrite(Num(a) + Num(b)).to(Num(a + b)) # Pushdown additon
yield rewrite(fib(a)).to(fib(a - 1) + fib(a - 2), a > 1) # Define fib recursively
yield rewrite(fib(a)).to(Num(a), a <= 1) # Define base cases
- create some rewrite rules
- define it recursively, and based on base cases
# 3. Add expresions of interest
egraph.register(fib(4))
# 4. Run rules
egraph.saturate()
- <Add to e-graph>
- there is only one expression in the graph
- outer circle shows equivalence class, currnetly its not
- As we saturate it, we see how it expands up
- extract out minimum cost
- extract out all versions
# 5. Extract out lowest cost equivalent expression
egraph.extract(fib(4))
Num(3)
egraph.extract_multiple(fib(4), 10)
[Num(3), Num(2) + Num(1), fib(4)]
Procedure for optimization
build types & functions to describe API
add rewrite rules
add expresions to graph
run rules
extract out lowest cost
Larger real world example#
from sklearn import config_context
from sklearn.datasets import make_classification
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# Binary classification with scikit-learn
def run_lda(x, y):
with config_context(array_api_dispatch=True):
lda = LinearDiscriminantAnalysis()
return lda.fit(x, y).transform(x)
X_np, y_np = make_classification(random_state=0, n_samples=1000000)
run_lda(X_np, y_np)
array([[ 0.64233002],
[ 0.63661245],
[-1.603293 ],
...,
[-1.1506433 ],
[ 0.71687176],
[-1.51119579]])
NumPy Array example
Build up symbolic array
arange 10 then get shape
concrete more practical example
lets say you are using scikit learn for in classification
you call funtion a lot with array, but its slow
What if we could partially evaluate it to create optimized form?
use this with numba to JIT compile to LLVM
totally omit the interfacing with Python interpreter
but numba cant optimize scikit learn, it doesnt know this dilect
but it does know numpy
from egglog.exp.array_api import *
x = NDArray.var("x")
x * 2
NDArray.var("x") * NDArray.scalar(Value.int(Int(2)))
from egglog.exp.array_api_jit import jit
@jit
def optimized_fn(X, y):
# Add metadata about input shapes and dtypes, so that abstract array
# can pass scikit-learn runtime checks
assume_dtype(X, X_np.dtype)
assume_shape(X, X_np.shape)
assume_isfinite(X)
assume_dtype(y, y_np.dtype)
assume_shape(y, y_np.shape)
assume_value_one_of(y, (0, 1))
return run_lda(X, y)
- By using egglog, we can translate between scikit learn domain and numpy
- if scikit-learn was written in egglog, this could be written as a re-write rule
- but its not, so what do we do?
- well because our array API conforms to the standards, we can pass in variable arrays
- shows power of egglog to work in environments where traditional python objects are expected
- we can call it
- we get back this big expression
- strongly typed
- mathematical
- in the dialect of this array api
- need to do a few translates actually to go into numba
- add rewrite rule to from mean to sum
- Then another one to go into Python string
- everything has been de-duplicated
- shape calculations have been partially evaluated
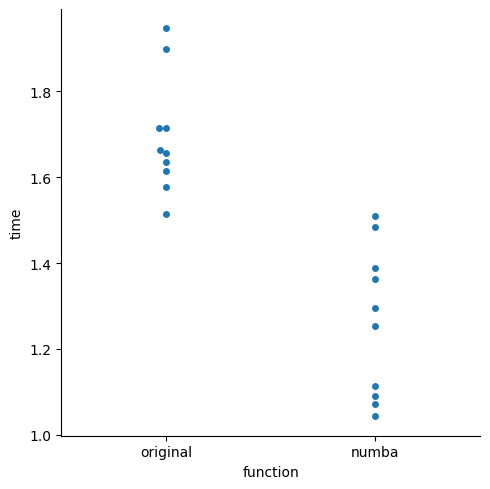
- if we evaluate peformance, we can see yes it did improve
optimized_fn.expr
_NDArray_1 = NDArray.var("X")
assume_dtype(_NDArray_1, DType.float64)
assume_shape(_NDArray_1, TupleInt(Int(1000000)) + TupleInt(Int(20)))
assume_isfinite(_NDArray_1)
_NDArray_2 = NDArray.var("y")
assume_dtype(_NDArray_2, DType.int64)
assume_shape(_NDArray_2, TupleInt(Int(1000000)))
assume_value_one_of(_NDArray_2, TupleValue(Value.int(Int(0))) + TupleValue(Value.int(Int(1))))
_NDArray_3 = astype(
NDArray.vector(TupleValue(sum(_NDArray_2 == NDArray.scalar(Value.int(Int(0)))).to_value()) + TupleValue(sum(_NDArray_2 == NDArray.scalar(Value.int(Int(1)))).to_value())),
DType.float64,
) / NDArray.scalar(Value.float(Float(1000000.0)))
_NDArray_4 = zeros(TupleInt(Int(2)) + TupleInt(Int(20)), OptionalDType.some(DType.float64), OptionalDevice.some(_NDArray_1.device))
_MultiAxisIndexKey_1 = MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(Slice()))
_IndexKey_1 = IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.int(Int(0))) + _MultiAxisIndexKey_1)
_NDArray_5 = _NDArray_1[ndarray_index(_NDArray_2 == NDArray.scalar(Value.int(Int(0))))]
_OptionalIntOrTuple_1 = OptionalIntOrTuple.some(IntOrTuple.int(Int(0)))
_NDArray_4[_IndexKey_1] = sum(_NDArray_5, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_5.shape[Int(0)]))
_IndexKey_2 = IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.int(Int(1))) + _MultiAxisIndexKey_1)
_NDArray_6 = _NDArray_1[ndarray_index(_NDArray_2 == NDArray.scalar(Value.int(Int(1))))]
_NDArray_4[_IndexKey_2] = sum(_NDArray_6, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_6.shape[Int(0)]))
_NDArray_7 = concat(TupleNDArray(_NDArray_5 - _NDArray_4[_IndexKey_1]) + TupleNDArray(_NDArray_6 - _NDArray_4[_IndexKey_2]), OptionalInt.some(Int(0)))
_NDArray_8 = square(_NDArray_7 - expand_dims(sum(_NDArray_7, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_7.shape[Int(0)]))))
_NDArray_9 = sqrt(sum(_NDArray_8, _OptionalIntOrTuple_1) / NDArray.scalar(Value.int(_NDArray_8.shape[Int(0)])))
_NDArray_10 = copy(_NDArray_9)
_NDArray_10[ndarray_index(_NDArray_9 == NDArray.scalar(Value.int(Int(0))))] = NDArray.scalar(Value.float(Float(1.0)))
_TupleNDArray_1 = svd(sqrt(NDArray.scalar(Value.float(Float.rational(Rational(1, 999998))))) * (_NDArray_7 / _NDArray_10), FALSE)
_Slice_1 = Slice(OptionalInt.none, OptionalInt.some(sum(astype(_TupleNDArray_1[Int(1)] > NDArray.scalar(Value.float(Float(0.0001))), DType.int32)).to_value().to_int))
_NDArray_11 = (_TupleNDArray_1[Int(2)][IndexKey.multi_axis(MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(_Slice_1)) + _MultiAxisIndexKey_1)] / _NDArray_10).T / _TupleNDArray_1[
Int(1)
][IndexKey.slice(_Slice_1)]
_TupleNDArray_2 = svd(
(sqrt((NDArray.scalar(Value.int(Int(1000000))) * _NDArray_3) * NDArray.scalar(Value.float(Float(1.0)))) * (_NDArray_4 - (_NDArray_3 @ _NDArray_4)).T).T @ _NDArray_11, FALSE
)
(
(_NDArray_1 - (_NDArray_3 @ _NDArray_4))
@ (
_NDArray_11
@ _TupleNDArray_2[Int(2)].T[
IndexKey.multi_axis(
_MultiAxisIndexKey_1
+ MultiAxisIndexKey(
MultiAxisIndexKeyItem.slice(
Slice(
OptionalInt.none,
OptionalInt.some(
sum(astype(_TupleNDArray_2[Int(1)] > (NDArray.scalar(Value.float(Float(0.0001))) * _TupleNDArray_2[Int(1)][IndexKey.int(Int(0))]), DType.int32))
.to_value()
.to_int
),
)
)
)
)
]
)
)[IndexKey.multi_axis(_MultiAxisIndexKey_1 + MultiAxisIndexKey(MultiAxisIndexKeyItem.slice(Slice(OptionalInt.none, OptionalInt.some(Int(1))))))]
import inspect
print(inspect.getsource(optimized_fn))
def __fn(X, y):
assert X.dtype == np.dtype(np.float64)
assert X.shape == (1000000, 20,)
assert np.all(np.isfinite(X))
assert y.dtype == np.dtype(np.int64)
assert y.shape == (1000000,)
assert set(np.unique(y)) == set((0, 1,))
_0 = y == np.array(0)
_1 = np.sum(_0)
_2 = y == np.array(1)
_3 = np.sum(_2)
_4 = np.array((_1, _3,)).astype(np.dtype(np.float64))
_5 = _4 / np.array(1000000.0)
_6 = np.zeros((2, 20,), dtype=np.dtype(np.float64))
_7 = np.sum(X[_0], axis=0)
_8 = _7 / np.array(X[_0].shape[0])
_6[0, :] = _8
_9 = np.sum(X[_2], axis=0)
_10 = _9 / np.array(X[_2].shape[0])
_6[1, :] = _10
_11 = _5 @ _6
_12 = X - _11
_13 = np.sqrt(np.array(float(1 / 999998)))
_14 = X[_0] - _6[0, :]
_15 = X[_2] - _6[1, :]
_16 = np.concatenate((_14, _15,), axis=0)
_17 = np.sum(_16, axis=0)
_18 = _17 / np.array(_16.shape[0])
_19 = np.expand_dims(_18, 0)
_20 = _16 - _19
_21 = np.square(_20)
_22 = np.sum(_21, axis=0)
_23 = _22 / np.array(_21.shape[0])
_24 = np.sqrt(_23)
_25 = _24 == np.array(0)
_24[_25] = np.array(1.0)
_26 = _16 / _24
_27 = _13 * _26
_28 = np.linalg.svd(_27, full_matrices=False)
_29 = _28[1] > np.array(0.0001)
_30 = _29.astype(np.dtype(np.int32))
_31 = np.sum(_30)
_32 = _28[2][:_31, :] / _24
_33 = _32.T / _28[1][:_31]
_34 = np.array(1000000) * _5
_35 = _34 * np.array(1.0)
_36 = np.sqrt(_35)
_37 = _6 - _11
_38 = _36 * _37.T
_39 = _38.T @ _33
_40 = np.linalg.svd(_39, full_matrices=False)
_41 = np.array(0.0001) * _40[1][0]
_42 = _40[1] > _41
_43 = _42.astype(np.dtype(np.int32))
_44 = np.sum(_43)
_45 = _33 @ _40[2].T[:, :_44]
_46 = _12 @ _45
return _46[:, :1]
import numba
import numpy as np
numba_fn = numba.njit(fastmath=True)(optimized_fn)
assert np.allclose(run_lda(X_np, y_np), numba_fn(X_np, y_np))
/var/folders/xn/05ktz3056kqd9n8frgd6236h0000gn/T/egglog-9befda2c-765c-4e2c-94d5-f667be45c7eb.py:56: NumbaPerformanceWarning: '@' is faster on contiguous arrays, called on (Array(float64, 2, 'C', False, aligned=True), Array(float64, 2, 'A', False, aligned=True))
_45 = _33 @ _40[2].T[:, :_44]
import timeit
import pandas as pd
import seaborn as sns
stmts = {
"original": "run_lda(X_np, y_np)",
"numba": "numba_fn(X_np, y_np)",
}
df = pd.DataFrame.from_dict(
{name: timeit.repeat(stmt, globals=globals(), number=1, repeat=10) for name, stmt in stmts.items()}
)
df_melt = pd.melt(df, var_name="function", value_name="time")
_ = sns.catplot(data=df_melt, x="function", y="time", kind="swarm")
What kind of analysis?#
egraph = EGraph([array_api_module])
_NDArray_1 = NDArray.var("X")
assume_dtype(_NDArray_1, DType.float64)
egraph.register(DType.float64 == _NDArray_1.dtype)
egraph.saturate()
assume_shape(_NDArray_1, TupleInt(Int(1000000)))
egraph = EGraph([array_api_module])
egraph.register(reshape(_NDArray_1, (-1,)).ndim == Int(1))
egraph.saturate()
# rule(
# greater_zero(v),
# greater_zero(v1),
# eq(v2).to(v / v1),
# ).then(
# greater_zero(v2),
# )
egraph = EGraph([array_api_module])
egraph.register( # Interval analysis
any((unique_counts(NDArray.var("X"))[1] / 1000000.0) < 0)
)
egraph.saturate()
Egglog can open a door to PL in Python#
Meet users where they are at
Introduce new abstractions to improve performance and interoperability between libraries
So whats the point?
all this mechanisms basically let us open a door
to do symbolic evaluate
bring formal methods into Python
If this can be a way to get research into the field
people arent going to use haskell, not write in coq,
they wont even use try a new python library
but, if all it takes is an import, and their code will go faster?
or they can target new hardware
thats compelling
Future work#
Can we implement this journey in egglog?
Python bytecode
-> RSVDG
-> abstract analysis of program state/effects ~Interaction Trees
-> Functional domains (a Mathematics of Arrays/APL)
<- Flatten arrays
<- Polyhedral forms/loop nests
GPU/LLVM code
Can this abstract interpretation, using a System F type system, and rewrite rules, serve as a way to communicate between these different domains?
What about studying the correctness properties of these rewrite rules and semantics?
Lenore mullin APL
functional representation of arrays
TODO quote from python mailing lists?
For example, what if we move the bytecode analysis numba does into this frame?
imperative -> functional code
RSVDG - start to use this work
maybe we could also use different models, from IT
take imperative code, lift into form to analyze the semantics, and optimize it
closer to a semantics
Let us express these different more abstract semantic domains, and go back and forth between those and imperative levels, of Python, of Array APIs
IF/when its working I would love to start exploring quesitons on the meta framework - like is the Systme F type system enough to represent all of this? - How can we prove properties of our system based on the rewrite rules?
Conclusion#
Python open source ecosystem needs more precise abstractions for building ecosystem of DSLs
Space for PL research to have a huge impact and lots of interesting mathematical problems
Thank you!
Saul Shanabrook
Before we can even begin to experiment here, we have to build up all this machinery
there is no compiler pass to add to Python
Either we have to fork the language, or do it through EDSLS
the latter seems more diverse, more fun, more collaborative
share horizontally
In a way, it’s a political question, that if we can add these ways to creating abstraction in Python, allows different ways of orienting social groups
work with numba team to use egglog to allow third party authors to add their own abstractions to the compiler that are erased
democratizing a compiler
rewrite rules that are much more readable than many other libraries
very mathametical
open up space for more people
This is a way of bringing PL research into high impact field
if we are to deliver here we also need to show it can work
I am applying for PhD programs to help this collaboration
I am familair with PyData ecosystem
need egglog to work
open research wquestions
need optimizations to work, need to have the right abstractions to store these things in a functional form
theory needs to hold up its of the bargain







