{post}
:author: Saul
egglog in Python¶
In this talk I will try to cover how egglog in Python can:
- Enable decentralized collaboration in the Python data science ecosystem.
- Provide a faithful authoring environment for
egglog.
Saul Shanabrook @ EGRAPHS Workshop - PLDI '23
Open Source Data Science Ecosystem in Python¶
The term “ecosystem” is often used to describe the modern open-source scientific software. In biology, the term “ecosystem” is defined as a biological community of interacting organisms and their physical environment. Modern open-source scientific software development occurs in a similarly interconnected and interoperable fashion.
from Jupyter Meets the Earth: Ecosystem
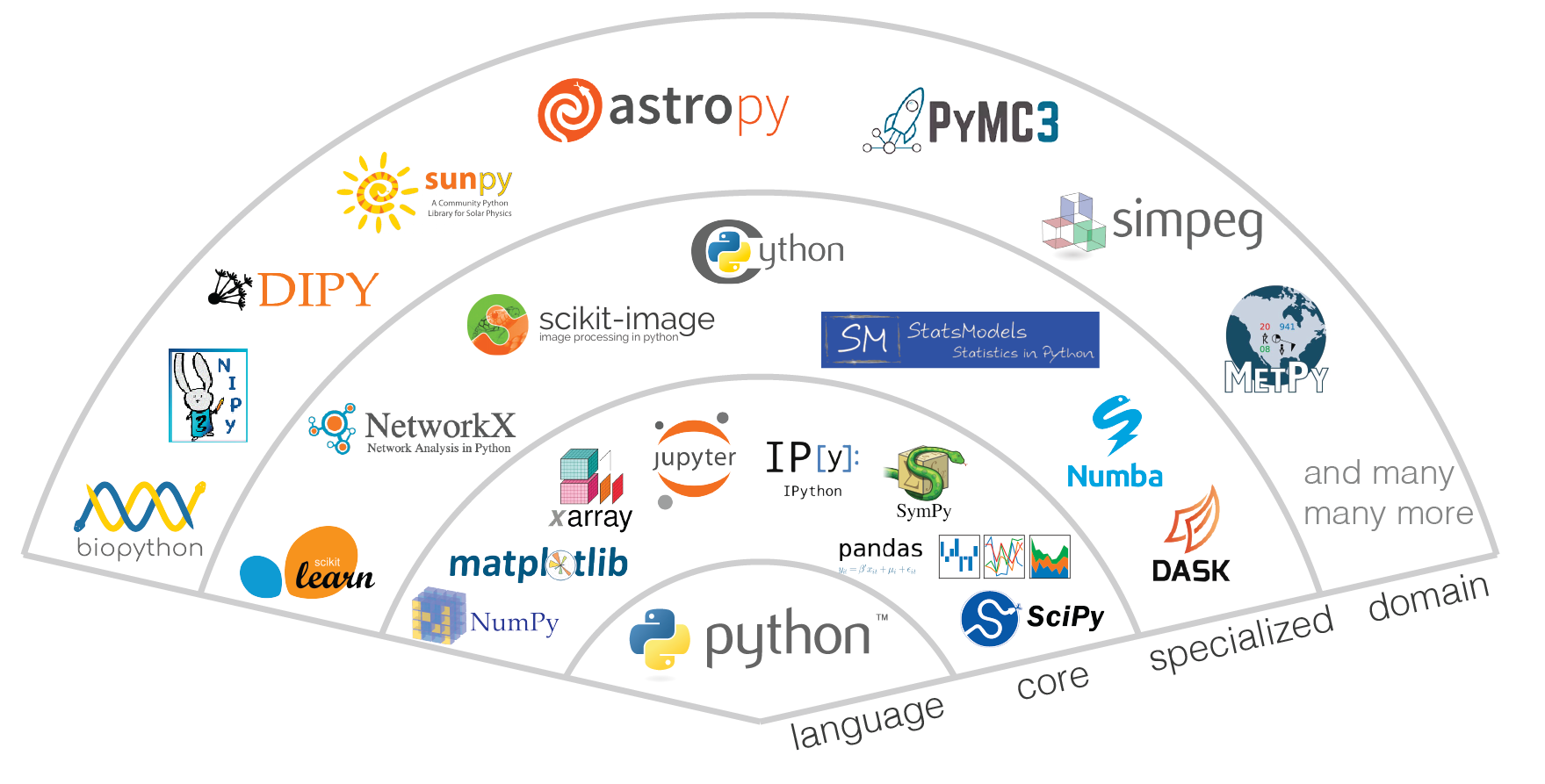
Aims¶
- How can the tools we build foster greater resiliancy, collaboration, and interdependence in this ecosystem?
- How can they help it stay flexible enough to adapt to the changing computational landscape to empower users and authors?
What role could egglog play?¶
- Bring the PL community closer to this space, providing theoretical frameworks for thinking about composition and language.
- Constrained type system could support decentralized interopability and composition between data science libraries.
Other Python EGraph Libraries¶
riswords/quiche: Optimizing Python ASTsegraphs-good/snake-egg: Generic bindings foregg, can bring any Python objects as expressions.- EGraph library added to
ibis: Conversions between dataframe syntax and SQL dialects
A story about Arrays¶
1. Someone makes an NDArray library...¶
In [32]:
ndarray_mod = Module()
...
In [34]:
@ndarray_mod.class_
class NDArray(Expr):
def __getitem__(self, idx: Values) -> Value:
...
def shape(self) -> Values:
...
@ndarray_mod.function
def arange(n: Value) -> NDArray:
...
Restifo Mullin, Lenore Marie, "A mathematics of arrays" (1988). Electrical Engineering and Computer Science - Dissertations. 249.
In [35]:
@ndarray_mod.register
def _(n: Value, idx: Values, a: NDArray):
yield rewrite(arange(n).shape()).to(Values(Vec(n)))
yield rewrite(arange(n)[idx]).to(idx[Value(0)])
In [38]:
egraph = EGraph([ndarray_mod])
ten = egraph.let("ten", arange(Value(10)))
ten_shape = ten.shape()
egraph.register(ten_shape)
egraph.run(20)
egraph.display()
egraph.extract(ten_shape)
Out[38]:
Values(Vec.empty().push(Value(10)))
In [41]:
ten_indexed = ten[Values(Vec(Value(7)))]
egraph.register(ten_indexed)
egraph.run(20)
egraph.display()
egraph.extract(ten_indexed)
Out[41]:
Value(7)
2. Someone else decides to implement a cross product library¶
In [21]:
cross_mod = Module([ndarray_mod])
@cross_mod.function
def cross(l: NDArray, r: NDArray) -> NDArray:
...
@cross_mod.register
def _cross(l: NDArray, r: NDArray, idx: Values):
yield rewrite(cross(l, r).shape()).to(l.shape().concat(r.shape()))
yield rewrite(cross(l, r)[idx]).to(l[idx] * r[idx])
In [22]:
egraph = EGraph([cross_mod])
egraph.simplify(cross(arange(Value(10)), arange(Value(11))).shape(), 10)
Out[22]:
Values(Vec.empty().push(Value(11)).push(Value(10)))
3. I write my wonderful data science application using it¶
In [23]:
def my_special_app(x: Value) -> Value:
return cross(arange(x), arange(x))[Values(Vec(x))]
egraph = EGraph([cross_mod])
egraph.simplify(my_special_app(Value(10)), 10)
Out[23]:
Value(100)
.... but its too slow...
In [24]:
for i in range(100):
egraph.simplify(my_special_app(Value(i)), 10)
4. Someone else writes a library for delayed execution¶
In [25]:
py_mod = Module([ndarray_mod])
@py_mod.function
def py_value(s: StringLike) -> Value:
...
...
Out[25]:
Ellipsis
5. I can use it jit compile my application!¶
In [27]:
egraph = EGraph([cross_mod, py_mod])
egraph.simplify(my_special_app(py_value("x")), 10)
Out[27]:
py_value("x * x")
Takeaways...¶
...from this totally realistic example.
- Declerative nature of
egglogcould facilitate decantralized library collaboration and experimentation.- Focus on types over values for library authors encourages interoperability.
- Pushing power down, empowering users and library authors
- Could allow greater collaboration between PL community and data science library community in Python
How does it work?¶
Sorts, expressions, and functions¶
In [3]:
%%egglog graph
(datatype Math
(Num i64)
(Var String)
(Add Math Math)
(Mul Math Math))
(define expr1 (Mul (Num 2) (Add (Var "x") (Num 3))))
(define expr2 (Add (Num 6) (Mul (Num 2) (Var "x"))))
Out[3]:
In [4]:
egraph = EGraph()
@egraph.class_
class Num(Expr):
@classmethod
def var(cls, name: StringLike) -> Num:
...
def __init__(self, value: i64Like) -> None:
...
def __add__(self, other: Num) -> Num:
...
def __mul__(self, other: Num) -> Num:
...
expr1 = egraph.let("expr1", Num(2) * (Num.var("x") + Num(3)))
expr2 = egraph.let("expr2", Num(6) + Num(2) * Num.var("x"))
egraph
Out[4]:
Rewrite rules and checks¶
In [5]:
%%egglog graph continue
(rewrite (Add a b)
(Add b a))
(rewrite (Mul a (Add b c))
(Add (Mul a b) (Mul a c)))
(rewrite (Add (Num a) (Num b))
(Num (+ a b)))
(rewrite (Mul (Num a) (Num b))
(Num (* a b)))
(run 10)
(check (= expr1 expr2))
Out[5]:
In [6]:
@egraph.register
def _(a: Num, b: Num, c: Num, i: i64, j: i64):
yield rewrite(a + b).to(b + a)
yield rewrite(a * (b + c)).to((a * b) + (a * c))
yield rewrite(Num(i) + Num(j)).to(Num(i + j))
yield rewrite(Num(i) * Num(j)).to(Num(i * j))
egraph.run(10)
egraph.check(eq(expr1).to(expr2))
egraph
Out[6]:
Extracting lowest cost expression¶
In [7]:
%%egglog continue output
(extract expr1)
Extracted with cost 8: (Add (Mul (Num 2) (Var "x")) (Num 6))
In [8]:
egraph.extract(expr1)
Out[8]:
(Num(2) * Num.var("x")) + Num(6)
Multipart Rules¶
In [9]:
%%egglog graph
(function fib (i64) i64)
(set (fib 0) 0)
(set (fib 1) 1)
(rule ((= f0 (fib x))
(= f1 (fib (+ x 1))))
((set (fib (+ x 2)) (+ f0 f1))))
(run 7)
(check (= (fib 7) 13))
Out[9]:
In [10]:
fib_egraph = EGraph()
@fib_egraph.function
def fib(x: i64Like) -> i64:
...
@fib_egraph.register
def _(f0: i64, f1: i64, x: i64):
yield set_(fib(0)).to(i64(1))
yield set_(fib(1)).to(i64(1))
yield rule(
eq(f0).to(fib(x)),
eq(f1).to(fib(x + 1)),
).then(set_(fib(x + 2)).to(f0 + f1))
fib_egraph.run(7)
fib_egraph.check(eq(fib(7)).to(i64(21)))
Include & Modules¶
In [11]:
%%writefile path.egg
(relation path (i64 i64))
(relation edge (i64 i64))
(rule ((edge x y))
((path x y)))
(rule ((path x y) (edge y z))
((path x z)))
Overwriting path.egg
In [12]:
%%egglog
(include "path.egg")
(edge 1 2)
(edge 2 3)
(edge 3 4)
(run 3)
(check (path 1 3))
In [13]:
mod = Module()
path = mod.relation("path", i64, i64)
edge = mod.relation("edge", i64, i64)
@mod.register
def _(x: i64, y: i64, z: i64):
yield rule(edge(x, y)).then(path(x, y))
yield rule(path(x, y), edge(y, z)).then(path(x, z))
In [14]:
egraph = EGraph([mod])
egraph.register(edge(1, 2), edge(2, 3), edge(3, 4))
egraph.run(3)
egraph.check(path(1, 3))
Possible next steps?¶
- Try getting toehold in existing library (like Ibis) to see if constrained egglog approach can still be useful.
- Add support for Python objects as builtin sort.
- Upstream egglog improvements which could help with reuse
- First class functions (would help with implementing things like reductions, mapping)
- User defined generic sorts (i.e. an array type agnostic to inner values)
Thank you!¶
pip install egglog
from egglog import *
egraph = EGraph()
...
Come say hello at github.com/egraphs-good/egglog-python!